Development of SNP array in Cherry, Peach, and Strawberry
- Umesh Rosyara
- Jun 6, 2012
- 2 min read
Rosaceous crops include fruit crops that are very important in human health due to high fruit value. Cheery and Peach belong to Prunus genus. Within Prunus (Rosaceae), two cherry species, sweet (P. avium) and sour cherry (P. cerasus). The sour cherry is tetraploid (2n = 4x = 32) and sweet cherry is diploid (2n = 2x = 16). Another Prunus member peach (Prunus persica) is diploid (2n = 2x = 16). Refrence genome is available for peach but not for cherries. However, extensive synteny conserved among the diploid Prunus species allowed us to use the peach genome as refrence point.

For SNP discovery purpose, in peach 56 peach breeding-relevant accessions while in cherry 16 sweet and 8 sour cherry accessions were subjected to low-depth genome re-sequencing . The reads were assembed to refrence genome of peach doubled haploid cultivar ‘Lovell’.
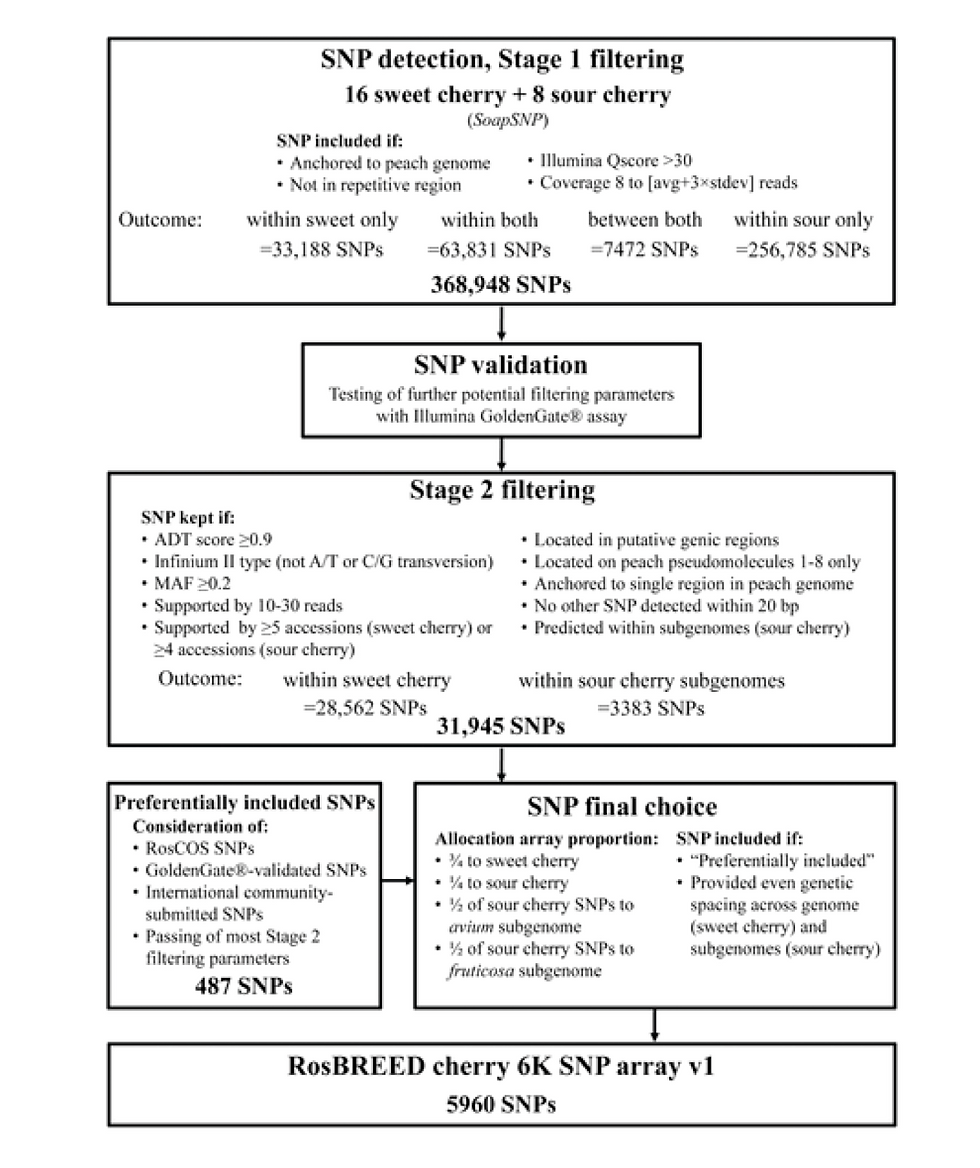
Example workflow of SNP array development
The cultivated strawberry, Fragaria × ananassa (Duch.), is an octoploid (assumed to be allopolyploid) (2n = 8x = 56) with no difinite genome model. Due process the Short-read sequences from one diploid and 19 octoploid accessions were aligned to the diploid Fragaria vesca ‘Hawaii 4’ reference genome. The resulting product is 90 K Affymetrix® Axiom® array consisting of single nucleotide polymorphisms (SNPs) and indels. Polyploidy makes the computation process much harders than in diploids. For this reason large number of markers were included in the array. Strawberry array is affymetrix array while cherry and peach arrray is Illunima arrray.
Publications:
Bassil N.V., ., U. R. Rosyara et al. 2015. Development and preliminary evaluation of a 90K Axiom SNP array for the Allo-octoploid cultivated strawberry Fragaria x ananassa}, BMC Genomics 2015, 16:155 doi:10.1186/s12864-015-1310-1
Verde I., N. Bassil, S. Scalabrin, B. Gilmore, C. T. Lawley, K. Gasic, D. Micheletti, U.R. Rosyara, F. Cattonaro, E. Vendramin, D. Main, V. Aramini, A.L. Blas, T.C. Mockler, D. W. Bryant, L.Wilhelm, M. Troggio, B. Sosinski, M. J. Aranzana, P.Arús, A. Iezzoni, M. Morgante, and C. Peace (2012) Development and evaluation of a 9K SNP array for peach by internationally coordinated SNP detection and validation in breeding germplasm, PLoS ONE 7(4): e35668. doi:10.1371/journal.pone.0035668
Peace C., D. Main, S. Ficklin, N. Bassil, B.Gilmore, U.R. Rosyara, A. Iezzoni, T. Stegmeir, A.Sebolt, C. Lawley, T. Mockler, and L.Wilhelm (2012) Genome-wide SNP detection, validation, and development of a 6K SNP assay for sweet cherry and sour cherry, PLoS ONE 7(12): e48305.
doi:10.1371/journal.pone.0048305
See also:
Comments